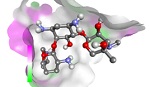
In Silico Analysis of Antibacterial Activity of Fatty Acids in Swietenia humilis Zucc. Seed Extract Against Staphylococcus aureus sortase A enzyme
Abstract
Keywords
Full Text:
PDFReferences
[1] M. Payne et al., "Synthesis and biological evaluation of tetrahydroisoquinoline-derived antibacterial compounds," Bioorganic & Medicinal Chemistry, vol. 57, p. 116648, 2022.
doi: 10.1016/j.bmc.2022.116648.
[2] T. J. Foster, "Antibiotic resistance in Staphylococcus aureus. Current status and future prospects," FEMS microbiology reviews, vol. 41, no. 3, pp. 430-449, 2017.
doi: 10.1093/femsre/fux007.
[3] Y. Zhao, J. Wei, C. Li, A. F. Ahmed, Z. Liu, and C. Ma, "A comprehensive review on mechanism of natural products against Staphylococcus aureus," Journal of Future Foods, vol. 2, no. 1, pp. 25-33, 2022.
doi: 10.1016/j.jfutfo.2022.03.014.
[4] H. Yu, M. Liu, Y. Liu, L. Qin, M. Jin, and Z. Wang, "Antimicrobial activity and mechanism of action of Dracocephalum moldavica L. extracts against clinical isolates of Staphylococcus aureus," Frontiers in Microbiology, vol. 10, p. 1249, 2019. doi: 10.3389/fmicb.2019.01249.
[5] S. Sasidharan, B. Prema, and L. Y. Latha, "Antimicrobial drug resistance of Staphylococcus aureus in dairy products," Asian Pacific Journal of Tropical Biomedicine, vol. 1, no. 2, pp. 130-132, 2011.
doi: 10.1016/S2221-1691(11)60010-5.
[6] A. S. Bayer, T. Schneider, and H. G. Sahl, "Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall," Annals of the New York Academy of Sciences, vol. 1277, no. 1, pp. 139-158, 2013.
doi:10.1111/j.17496632.2012.06819.x.
[7] A. Pantosti, A. Sanchini, and M. Monaco, "Mechanisms of antibiotic resistance in Staphylococcus aureus," Future Microbio, vol. 2, no. 3, pp. 323, 2007.
doi: 10.2217/17460913.2.3.323.
[8] P. Jangra and A. Singh, "Staphylococcus aureus β-hemolysin-neutralizing single-domain antibody isolated from phage display library of Indian desert camel," Asian Pacific Journal of Tropical Medicine, vol. 3, no. 1, pp. 1-7, 2010.
doi:10.1016/S1995-7645(10)60020-X.
[9] H. Lade, H.S. Joo, and J.-S. Kim, "Molecular basis of non-β-lactam antibiotics resistance in Staphylococcus aureus," Antibiotics, vol. 11, no. 10, p. 1378, 2022.
doi: 10.3390/antibiotics11101378.
[10] L. M. Assis, M. Nedeljković, and A. Dessen, "New strategies for targeting and treatment of multi-drug resistant Staphylococcus aureus," Drug Resistance Updates, vol. 31, pp. 1-14, 2017.
doi: 10.1016/j.drup.2017.03.001.
[11] R. H. Baltz, "Daptomycin: mechanisms of action and resistance, and biosynthetic engineering," Current opinion in chemical biology, vol. 13, no. 2, pp. 144-151, 2009.
doi: 10.1016/j.cbpa.2009.02.031.
[12] A. Upadhyay, I. Upadhyaya, A. Kollanoor-Johny, and K. Venkitanarayanan, "Combating pathogenic microorganisms using plant‐derived antimicrobials: a minireview of the mechanistic basis," BioMed research international, vol. 2014, no. 1, p. 761741, 2014.
doi; 10.1155/2014/761741.
[13] A. G. Atanasov, S. B. Zotchev, V. M. Dirsch, and C. T. Supuran, "Natural products in drug discovery: advances and opportunities," Nature reviews Drug discovery, vol. 20, no. 3, pp. 200-216, 2021.
doi: 10.1038/s41573-020-00114-z.
[14] S. Mulyani, A. Kusumawardani, and A. A. Pangesti, "The Antibacterial Activity of Liquid Soap supplemented with Extracts combination of Cyperus rotundus L. and Flowers of Plumeria acuminata, Michelia alba, or Cananga odorata Against Staphylococcus aureus and Escherichia coli Bacteria," JKPK (Jurnal Kimia dan Pendidikan Kimia), vol. 7, no. 1, pp. 125-137, 2022.
doi: 10.20961/jkpk.v7i1.61033.
[15] S.-H. Mun et al., "Curcumin reverse methicillin resistance in Staphylococcus aureus," Molecules, vol. 19, no. 11, pp. 18283-18295, 2014.
doi: 10.3390/molecules191118283.
[16] M. W. Ha, S. W. Yi, and S.-M. Paek, "Design and synthesis of small molecules as potent staphylococcus aureus sortase a inhibitors," Antibiotics, vol. 9, no. 10, p. 706, 2020. doi: 10.3390/antibiotics9100706.
[17] S. Alharthi, S. E. Alavi, P. M. Moyle, and Z. M. Ziora, "Sortase A (SrtA) inhibitors as an alternative treatment for superbug infections," Drug Discovery Today, vol. 26, no. 9, pp. 2164-2172, 2021.
doi: 10.1016/j.drudis.2021.04.001.
[18] W. J. Bradshaw, A. H. Davies, C. J. Chambers, A. K. Roberts, C. C. Shone, and K. R. Acharya, "Molecular features of the sortase enzyme family," The FEBS journal, vol. 282, no. 11, pp. 2097-2114, 2015.
doi: 10.1111/febs.13282.
[19] L. Wang et al., "Eriodictyol as a potential candidate inhibitor of sortase A protects mice from methicillin-resistant Staphylococcus aureus-induced pneumonia," Frontiers in microbiology, vol. 12, p. 635710, 2021. doi: 10.3389/fmicb.2021.635710.
[20] L. A. Marraffini, A. C. DeDent, and O. Schneewind, "Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria," Microbiology and molecular biology reviews, vol. 70, no. 1, pp. 192-221, 2006.
doi:10.1128/MMBR.70.1.192221.2006.
[21] C. P. Gordon, P. Williams, and W. C. Chan, "Attenuating Staphylococcus aureus virulence gene regulation: a medicinal chemistry perspective," Journal of medicinal chemistry, vol. 56, no. 4, pp. 1389-1404, 2013.
doi: 10.1021/jm301745g.
[22] L. L. Zhou and C. G. Yang, "Chemical intervention on Staphylococcus aureus Virulence," Chinese Journal of Chemistry, vol. 37, no. 2, pp. 183-193, 2019.
doi: 10.1002/cjoc.201800470.
[23] Z. Taj and I. Chattopadhyay, "Staphylococcus aureus Virulence Factors and Biofilm Components: Synthesis, Structure, Function and Inhibitors," in ESKAPE Pathogens: Detection, Mechanisms and Treatment Strategies: Springer, 2024, pp. 227-270.
doi: 10.1007/978-981-99-8799-3_8.
[24] L. Tian, L. Wang, F. Yang, T. Zhou, and H. Jiang, "Exploring the modulatory impact of isosakuranetin on Staphylococcus aureus: Inhibition of sortase A activity and α-haemolysin expression," Virulence, vol. 14, no. 1, p. 2260675, 2023.
doi:10.1080/21505594.2023.2260675.
[25] E. P. Ivanova et al., "Bactericidal activity of self-assembled palmitic and stearic fatty acid crystals on highly ordered pyrolytic graphite," Acta Biomaterialia, vol. 59, pp. 148-157, 2017.
doi: 10.1016/j.actbio.2017.07.004.
[26] F. M. Joujou, N. E. Darra, H. N. Rajha, E. S. Sokhn, and N. Alwan, "Evaluation of synergistic/antagonistic antibacterial activities of fatty oils from apricot, date, grape, and black seeds," Scientific Reports, vol. 14, no. 1, p. 6532, 2024. doi: 10.1038/s41598-024-54850-y.
[27] S. K. Khadke, J.-H. Lee, Y.-G. Kim, V. Raj, and J. Lee, "Assessment of antibiofilm potencies of nervonic and oleic acid against Acinetobacter baumannii using in vitro and computational approaches," Biomedicines, vol. 9, no. 9, p. 1133, 2021.
doi: 10.3390/biomedicines9091133.
[28] G. Casillas-Vargas et al., "Antibacterial fatty acids: An update of possible mechanisms of action and implications in the development of the next-generation of antibacterial agents," Progress in lipid research, vol. 82, p. 101093, 2021.
doi: 10.1016/j.plipres.2021.101093.
[29] C. B. Huang, B. George, and J. L. Ebersole, "Antimicrobial activity of n-6, n-7 and n-9 fatty acids and their esters for oral microorganisms," Archives of oral biology, vol. 55, no. 8, pp. 555-560, 2010.
doi:10.1016/j.archoralbio.2010.06.007
[30] A. Asmara, C. Nuzlia, and R. Maryana, "Antibacterial bioactivity of n-hexane extract from Mahogany (Swietenia humilis Zucc.) seed and its fatty acid compound identification," in IOP Conference Series: Earth and Environmental Science, 2019, vol. 251, no. 1: IOP Publishing, p. 012016. doi:10.1088/1755-1315/251/1/012016.
[31] F. Stanzione, I. Giangreco, and J. C. Cole, "Use of molecular docking computational tools in drug discovery," Progress in Medicinal Chemistry, vol. 60, pp. 273-343, 2021. doi: 10.1016/bs.pmch.2021.01.005.
[32] J. Fan, A. Fu, and L. Zhang, "Progress in molecular docking," Quantitative Biology, vol. 7, pp. 83-89, 2019.
doi: 10.1007/s40484-019-0184-6.
[33] J. Setyono, S. C. Nurani, M. S. Fareza, A. Fadlan, and S. Sarmoko, "Molecular Docking of 6-shogaol and Curcumin on DNMT1 and LSD1 As Potential Agents for Thalassemia Treatment," JKPK (Jurnal Kimia dan Pendidikan Kimia), vol. 6, no. 3, pp. 326-334.
doi: 10.20961/jkpk.v6i3.40132.
[34] S. D. Oniga et al., "New 2-phenylthiazoles as potential sortase A inhibitors: Synthesis, biological evaluation and molecular docking," Molecules, vol. 22, no. 11, p. 1827, 2017.
doi: 10.3390/molecules22111827.
[35] M. A. Mahmood Janlou, H. Sahebjamee, M. Yazdani, and L. Fozouni, "Structure-based virtual screening and molecular dynamics approaches to identify new inhibitors of Staphylococcus aureus sortase A," Journal of Biomolecular Structure and Dynamics, vol. 42, no. 3, pp. 1157-1169, 2024.
doi: 10.1080/07391102.2022.203905.
[36] K. Liu et al., "The Discovery of Novel Agents against Staphylococcus aureus by Targeting Sortase A: A Combination of Virtual Screening and Experimental Validation," Pharmaceuticals, vol. 17, no. 1, p. 58, 2023.
doi: 10.3390/ph17010058
[37] R. Sapra, A. K. Rajora, P. Kumar, G. P. Maurya, N. Pant, and V. Haridas, "Chemical biology of sortase a inhibition: A gateway to anti-infective therapeutic agents," Journal of Medicinal Chemistry, vol. 64, no. 18, pp. 13097-13130, 2021.
doi: 10.1021/acs.jmedchem.1c00672
[38] I. PerkinElmer Informatics, ChemDraw 13.0. Waltham, MA, USA: PerkinElmer Informatics, Inc., 2013.
[39] I. Hypercube, HyperChem 8.0. Gainesville, FL, USA: Hypercube, Inc., 2007.
[40] M. Seeliger and B. de Groot, PyRx. La Jolla, CA, USA: The Scripps Research Institute, 2010.
[41] I. BioDiscovery, BioDiscovery Studio 12. El Segundo, CA, USA: BioDiscovery, Inc.
[42] T. O. B. D. Team, Open Babel 3.1.1. Open Babel Project, 2020.
[43] N. Suree et al., "The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized," Journal of Biological Chemistry, vol. 284, no. 36, pp. 24465-24477, 2009.
doi: 10.1074/jbc.M109.032151.
[44] J. J. Irwin and B. K. Shoichet. "ZINC8143541 (Gentamicin) " University of California, San Francisco. http://zinc15.docking.org/ (accessed 20 August, 2022).
[45] P. Varvarà, C. Calà, C. M. Maida, M. Giuffrè, N. Mauro, and G. Cavallaro, "Arginine-rich peptidomimetic ampicillin/gentamicin conjugate to tackle nosocomial biofilms: a promising strategy to repurpose first-line antibiotics," ACS Infectious Diseases, vol. 9, no. 4, pp. 916-927, 2023.
doi: 10.1021/acsinfecdis.2c00579.
[46] X. Hou, J. Du, J. Zhang, L. Du, H. Fang, and M. Li, "How to improve docking accuracy of AutoDock4. 2: a case study using different electrostatic potentials," Journal of Chemical Information and Modeling, vol. 53, no. 1, pp. 188-200, 2013.
doi: 10.1021/ci300417y.
[47] N. Alfisah, M. Masriany, and H. Hafsan, "Molecular Docking of Shallot (Allium ascalonicum) Active Compounds to Lanosterol Enzym 14-Alpha Demethylase and Squalene Monooxygenase for Antifungi Potential Activity," JKPK (Jurnal Kimia dan Pendidikan Kimia), vol. 7, no. 3, pp. 359-367.
doi: 10.20961/jkpk.v7i3.41510.
[48] B. Arsic, Y. Zhu, D. E. Heinrichs, and M. J. McGavin, "Induction of the staphylococcal proteolytic cascade by antimicrobial fatty acids in community acquired methicillin resistant Staphylococcus aureus," 2012.
doi: 10.1371/journal.pone.0045952.
[49] K. B. Tiwari, C. Gatto, and B. J. Wilkinson, "Plasticity of coagulase-negative staphylococcal membrane fatty acid composition and implications for responses to antimicrobial agents," Antibiotics, vol. 9, no. 5, p. 214, 2020. doi: 10.3390/antibiotics9050214.
[50] C. Subramanian, M. W. Frank, J. L. Batte, S. G. Whaley, and C. O. Rock, "Oleate hydratase from Staphylococcus aureus protects against palmitoleic acid, the major antimicrobial fatty acid produced by mammalian skin," Journal of Biological Chemistry, vol. 294, no. 23, pp. 9285-9294, 2019.
doi: 10.1074/jbc.RA119.008439.
[51] Y. Wu, Y. Sun, Z. Zhang, J. Chen, and G. Dong, "Effects of peptidoglycan, lipoteichoic acid and lipopolysaccharide on inflammation, proliferation and milk fat synthesis in bovine mammary epithelial cells," Toxins, vol. 12, no. 8, p. 497, 2020.
doi: 10.3390/toxins12080497.
[52] A. E. Sidders et al., "Antibiotic-induced accumulation of lipid II sensitizes bacteria to antimicrobial fatty acids," bioRxiv, p. 2022.05. 03.490474, 2022. doi: 10.1101/2022.05.03.490474.
[53] J. G. Kenny et al., "The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications," PLoS one, vol. 4, no. 2, p. e4344, 2009.
doi: 10.1371/journal.pone.0004344.
[54] A. Colautti, E. Orecchia, G. Comi, and L. Iacumin, "Lactobacilli, a weapon to counteract pathogens through the inhibition of their virulence factors," Journal of Bacteriology, vol. 204, no. 11, pp. e00272-22, 2022.
doi: 10.1128/jb.00272-22.
[55] R. Sangavi et al., "In silico analysis unravels the promising anticariogenic efficacy of fatty acids against dental caries causing Streptococcus mutans," Journal of Biomolecular Structure and Dynamics, pp. 1-16, 2023.
doi: 10.1080/07391102.2023.2211234
[56] K. Widmer, K. Soni, M. Hume, R. Beier, P. Jesudhasan, and S. Pillai, "Identification of poultry meat‐derived fatty acids functioning as quorum sensing signal inhibitors to autoinducer‐2 (AI‐2)," Journal of Food Science, vol. 72, no. 9, pp. M363-M368, 2007.
doi:10.1111/j.1750-3841.2007.00552.x
[57] S. R. Clarke et al., "The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin," Cell host & microbe, vol. 1, no. 3, pp. 199-212, 2007.
doi: 10.1016/j.chom.2007.04.005.
[58] K. R. V. Thappeta et al., "In-silico identified new natural sortase a inhibitors disrupt S. aureus biofilm formation," International Journal of Molecular Sciences, vol. 21, no. 22, p. 8601, 2020.
doi: 10.3390/ijms21228601.
[59] B.-X. Tian and L. A. Eriksson, "Catalytic mechanism and roles of Arg197 and Thr183 in the Staphylococcus aureus sortase A enzyme," The Journal of Physical Chemistry B, vol. 115, no. 44, pp. 13003-13011, 2011.
doi: 10.1021/jp207323y.
[60] B. A. Frankel, Y. Tong, M. L. Bentley, M. C. Fitzgerald, and D. G. McCafferty, "Mutational analysis of active site residues in the Staphylococcus aureus transpeptidase SrtA," Biochemistry, vol. 46, no. 24, pp. 7269-7278, 2007. doi: 10.1021/bi700100g.
Refbacks
- There are currently no refbacks.